| Log | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_log.txt |
| Query genes | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_query_genes.tab |
footprint-discovery -v 1 -nodie -org Escherichia_coli_GCF_000005845.2_ASM584v2 -taxon Gammaproteobacteria -genes $RSAT/public_html/tmp/apache/2018/03/21/footprint-discovery_Gammaproteobacteria_Escherichia_coli_GCF_000005845.2_ASM584v2_lexA_2018-03-21.142439_2018-03-21.142439_C9MNSJ/lexA_genes -sep_genes -lth occ 1 -lth occ_sig 0 -uth rank 50 -return occ,proba,rank -filter -bg_model taxfreq -task query_seq,filter_dyads,orthologs,ortho_seq,purge,dyads,maps,gene_index,index -o $RSAT/public_html/tmp/apache/2018/03/21/footprint-discovery_Gammaproteobacteria_Escherichia_coli_GCF_000005845.2_ASM584v2_lexA_2018-03-21.142439_2018-03-21.142439_C9MNSJ
Program version 1.116
Working directory /space23/rsat/public_html
Computer name sinik
Query organism Escherichia_coli_GCF_000005845.2_ASM584v2
Reference taxon Gammaproteobacteria
Query gene lexA
Background model taxfreq
Dyad filtering ON
| Query sequence | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_query_seq.fasta |
| Filter dyads | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_filter_dyads.tab |
| Orthologs | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_bbh.tab |
| ortho sequences | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_seq.fasta |
| Purged sequences | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_seq_purged.fasta |
| Dyads (tab format) | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_dyads_3nt_sp0-20-2str-noov_taxfreq_sig0.tab |
| Dyads (html format) | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_dyads_3nt_sp0-20-2str-noov_taxfreq_sig0.html |
| Assembled dyads | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_dyads_3nt_sp0-20-2str-noov_taxfreq_sig0.asmb |
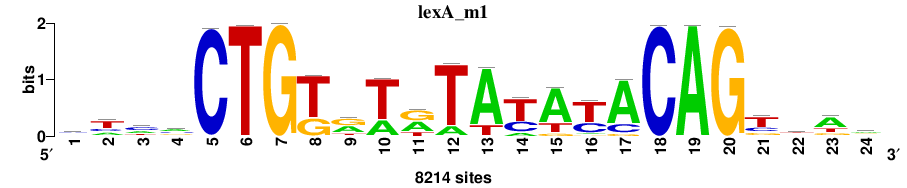
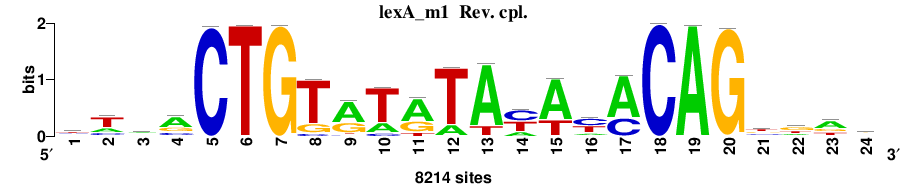
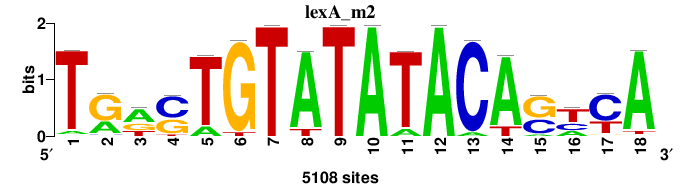
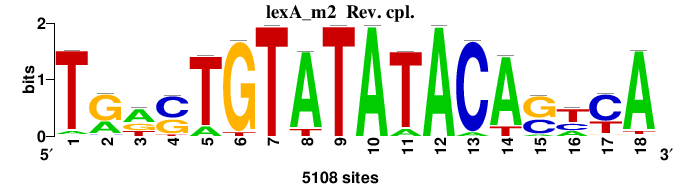
| Significance matrices (intermediate result) | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_dyads_3nt_sp0-20-2str-noov_taxfreq_sig0_pssm_sig_matrices.tf |
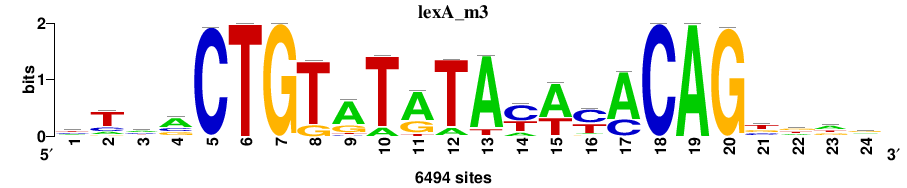
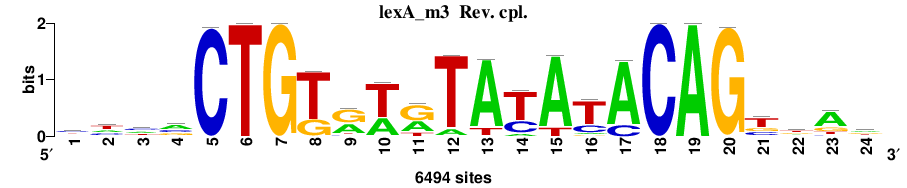
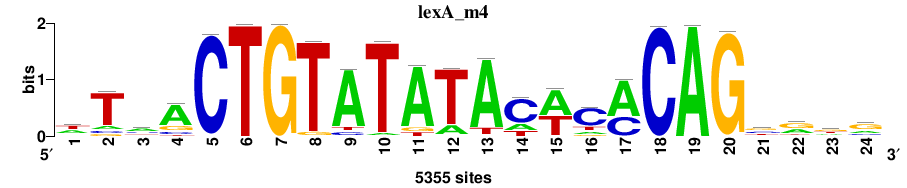
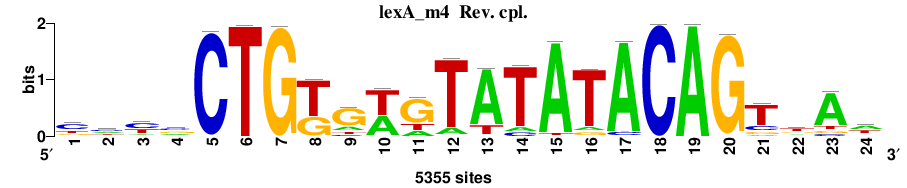
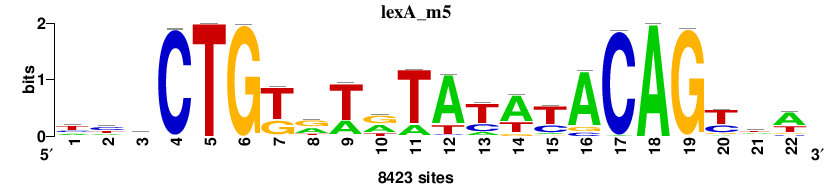
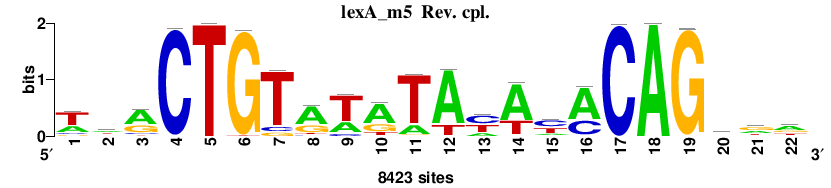
| Final matrices (tab format) | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_dyads_3nt_sp0-20-2str-noov_taxfreq_sig0_pssm_count_matrices.txt |
| Final matrices (transfac format) | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_dyads_3nt_sp0-20-2str-noov_taxfreq_sig0_pssm_count_matrices.tf |
| 5 matrices |
|
| Dyad map | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_dyads_3nt_sp0-20-2str-noov_taxfreq_sig0.png |
| (Click on image below) |
 |
| matrix-scan map | lexA_Escherichia_coli_GCF_000005845.2_ASM584v2_Gammaproteobacteria_taxfreq_ortho_dyads_3nt_sp0-20-2str-noov_taxfreq_sig0_pssm.png |
| (Click on image below) |
 |
Query status: