footprint-scan Gammaproteobacteria Escherichia_coli_GCF_000005845.2_ASM584v2 Matrix
Command: footprint-scan -v 0 -nodie -synthesis -sep_genes -org Escherichia_coli_GCF_000005845.2_ASM584v2 -taxon Gammaproteobacteria -m $RSAT/public_html/tmp/apache/2018/03/21/footprint-scan_Gammaproteobacteria__lexA_recA_2018-03-21.183943_2018-03-21.183943_J8t3qJ/input_matrix -matrix_format transfac -genes $RSAT/public_html/tmp/apache/2018/03/21/footprint-scan_Gammaproteobacteria__lexA_recA_2018-03-21.183943_2018-03-21.183943_J8t3qJ/lexA_recA_genes -info_lines -bginput -markov 0 -pval 1e-4 -occ_th 5 -plot_format jpg -map_format jpg -o $RSAT/public_html/tmp/apache/2018/03/21/footprint-scan_Gammaproteobacteria__lexA_recA_2018-03-21.183943_2018-03-21.183943_J8t3qJ/lexA_recA
| gene |
#matrix |
score |
occ |
occ_cum |
inv_cum |
inv_cum_freq |
occ_prior |
exp_occ |
occ_Pval |
occ_Eval |
occ_sig |
weight_at_th |
occ_at_th |
exp_at_th |
sig_at_th |
OCC_Sig |
SigPlot |
FreqPlot |
Map |
name |
upstr_neighb_name |
descr |
| recA |
LexA |
9.9 |
2 |
107812 |
274 |
2.535065e-03 |
5.522769e-06 |
0.60 |
0.000000e+00 |
0.000000e+00 |
350.00 |
6.0 |
2 |
10.47 |
350.00 | [sig] |
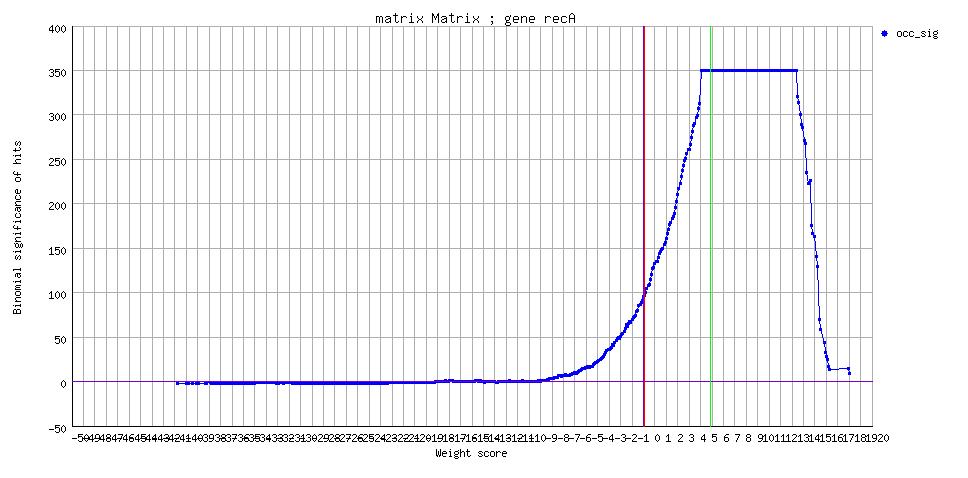 [sigplot] [sigplot] |
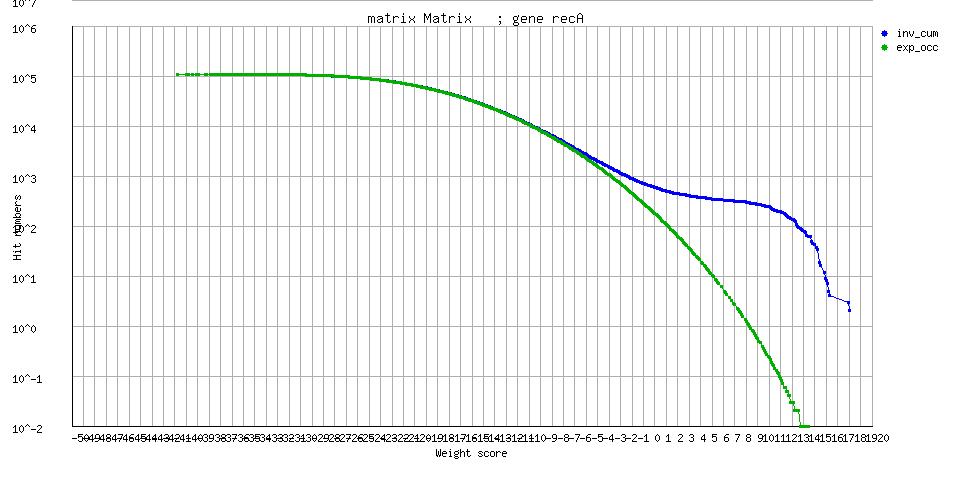 [freqplot] [freqplot] |
[map] |
recA |
NA |
NA |
| lexA |
LexA |
9.9 |
4 |
134724 |
316 |
2.340117e-03 |
5.193126e-06 |
0.70 |
0.000000e+00 |
0.000000e+00 |
350.00 |
5.9 |
4 |
12.98 |
350.00 | [sig] |
 [sigplot] [sigplot] |
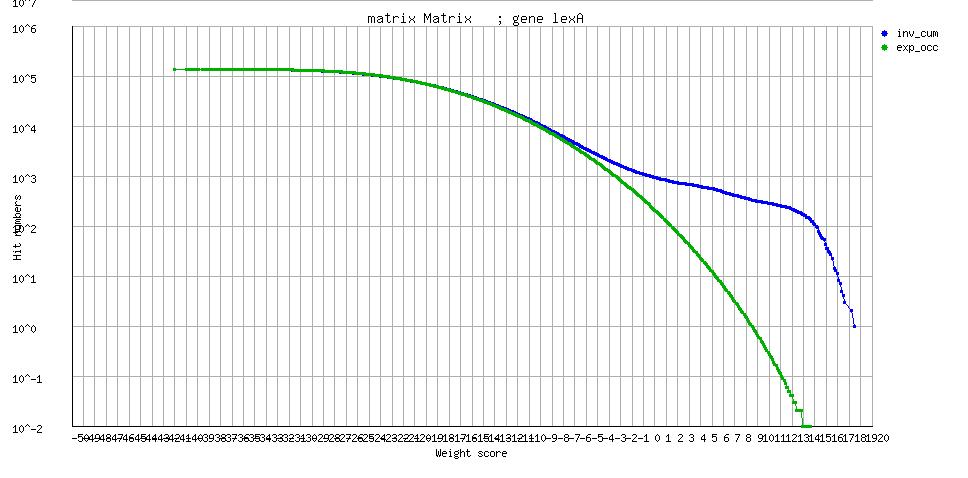 [freqplot] [freqplot] |
[map] |
lexA |
NA |
NA |
; Host name sinik
; Job started 2018-03-21.183944
; Job done 2018-03-21.185442
; Seconds 26.63
; user 26.63
; system 0.28
; cuser 801.87
; csystem 113.54
 [sigplot]
[sigplot]  [freqplot]
[freqplot]  [sigplot]
[sigplot]  [freqplot]
[freqplot]  [sigplot]
[sigplot]  [freqplot]
[freqplot]  [sigplot]
[sigplot]  [freqplot]
[freqplot]